All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MPN Advocates Network.
The mpn Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mpn Hub cannot guarantee the accuracy of translated content. The mpn and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The MPN Hub is an independent medical education platform, sponsored by AOP Health, GSK, Sumitomo Pharma, and supported through educational grants from Bristol Myers Squibb and Incyte. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MPN content recommended for you
Gene expression signature as a risk stratification tool for patients with myelofibrosis
Myelofibrosis (MF) is a complex hematologic disorder that develops due to certain mutations in hematopoietic stem cells. Primary and secondary MF (PMF and SMF, respectively) share common histopathologic features and clinical manifestations, and therefore, are currently managed in the same way.
Currently, a number of prognostic models are in use to predict survival outcomes for patients with MF such as International Prognostic Scoring System (IPSS) and dynamic IPSS (DIPSS) based on histopathologic features, mutation-enhanced IPSS (MIPSS70), and genetically-inspired IPSS (GIPSS). However, these models do not distinguish between the different subtypes such as pre-MF, PMF, and SMF despite clear differences in their course.
To provide more precision in the predicting outcome, Rontauroli et al. reported in Blood Advances, a correlation between gene expression profile (GEP) in MF subtypes and survival outcomes.1
The study aimed to identify whether GEP correlates with outcome (in general) and can provide more precision in predicting the risk of death or progression for PMF and SMF.
Methods
- JAK2V617F and MPLW515x mutations were identified via real-time quantitative PCR.
- CALR mutation was detected through capillary electrophoresis and bidirectional sequencing.
- High molecular risk mutations (HMR) such as ASXL1, EZH2, IDH1, IDH2, and SRFSF2 were identified through next-generation sequencing (NGS).
- The classification model was built starting from the list of probe sets resulting from Cox regression analysis and risk classes defined with hierarchical clustering.
- Probe set obtained from Cox regression analysis was used to create a gene expression-based classifier or the genetic expression model, and the ‘nearest shrunken centroids’ method was used to construct the model. Also, for 20-fold cross-validation strategy, pamr.cv R function was used.
Patient characteristics
Granulocyte samples from 114 patients with PMF/SMF were retrospectively analyzed according to clinical subtype including pre-PMF, overt PMF, post-essential thrombocytosis MF (PET-MF), and post- polycythemia vera MF (PPV-MF).
Table 1 includes information of patient characteristics with traditional risk scoring.
Table 1. Patient characteristics based on the diagnosis of MF clinical subtype.*
|
ASXL1, polycomb chromatin-binding protein gene; DIPSS, Dynamic IPSS; EZH2, enhancer of zeste homolog 2 gene; Hb, hemoglobin; HMR, high molecular risk; IDH, isocitrate dehydrogenase gene; Int, intermediate; IPSS, International Prognostic Scoring System; JAK2V617F, Janus kinase 2 mutation in phenylalanine at position 617; MF, myelofibrosis; MIPSS, mutation-enhanced IPSS; NA, not available; PET-MF, post-essential thrombocytosis MF; PMF, primary MF; PPV-MF, post-polycythemia vera MF; SRSF2, serine and arginine-rich splicing factor 2 gene. |
|||||
|
Variable |
Pre-PMF |
Overt PMF |
PET-MF |
PPV-MF |
p value† |
|---|---|---|---|---|---|
|
Median follow-up, years (range) |
6.88 |
5.54 |
4.55 |
4.18 |
6.15 × 10−1 |
|
Males, % |
54.3 |
56.8 |
50.0 |
43.8 |
8.33 × 10−1 |
|
Median age, years (range) |
62.90 |
63.80 |
65.80 |
71.10 |
9.62 × 10−2 |
|
Hb, median (range), g/dL |
12.40 |
11.20 |
10.75 |
12.55 |
2.52 × 10−4 |
|
Hb <10 g/dL, % |
5.7 |
24.3 |
34.6 |
12.5 |
2.75 × 10−2 |
|
Leukocytes, median (range), × 109/L |
8.70 (3.6−41.0) |
10.00 (2.8−89.0) |
9.58 (2.3−104.0) |
14.90 (5.9−88.7) |
1.35 × 10−2 |
|
Leukocytes >25 × 109/L, % |
8.8 |
16.2 |
11.5 |
46.7 |
9.09 × 10−3 |
|
Platelets, median (range), × 109/L |
410.0 (72−1299) |
179.0 (22−1252) |
377.5 (61−1568) |
224.5 (20−1271) |
6.54 × 10−3 |
|
Splenomegaly, % |
45.7 |
86.1 |
84.6 |
71.4 |
6.24 × 10−4 |
|
JAK2V617F, % |
12 |
16 |
11 |
15 |
8.04 × 10−4 |
|
ASXL1 mutation, % |
44.0 |
31.2 |
33.3 |
46.2 |
6.81 × 10−1 |
|
EZH2 mutation, % |
12.5 |
7.4 |
0 |
6.7 |
5.27 × 10−1 |
|
SRSF2 mutation, % |
30.4 |
3.7 |
6.2 |
0 |
5.98 × 10−3 |
|
IDH1/2 mutation, % |
8.7 |
6.7 |
0 |
6.7 |
6.36 × 10−1 |
|
HMR, % |
56.0 |
41.4 |
35.3 |
46.7 |
5.66 × 10−1 |
|
HMR ≥2, % |
32.0 |
6.9 |
0 |
6.7 |
6.82 × 10−3 |
|
DIPSS (n evaluable, |
31 |
36 |
25 |
15 |
— |
|
Low, % |
45.2 |
25.0 |
16.0 |
20.0 |
— |
|
Int-1, % |
32.3 |
36.1 |
52.0 |
33.3 |
— |
|
Int-2, % |
9.7 |
33.3 |
16.0 |
33.3 |
— |
|
High, % |
12.9 |
5.6 |
16.0 |
13.3 |
3.89 × 10−1 |
|
MIPSS70 (n evaluable, |
21 |
25 |
15 |
12 |
— |
|
Low, % |
47.6 |
8.0 |
0 |
8.3 |
— |
|
Intermediate, % |
19.0 |
56.0 |
66.7 |
41.7 |
— |
|
High, % |
33.3 |
36.0 |
33.3 |
50.0 |
1.71 × 10−3 |
|
Progression to leukemia, % |
22.9 |
8.1 |
7.7 |
0 |
6.14 × 10−2 |
|
Death, % |
37.1 |
45.9 |
34.6 |
62.5 |
2.78 × 10−1 |
Gene signature for hierarchal clustering and OS
Based on Cox-regression analysis, 596 genes were found to be correlated with survival, of which 433 were associated with inferior survival. According to the expression of these genes, a hierarchical clustering was performed. Subsequently, patient samples were reclassified into a high and low-risk group. Overall survival (OS) for the lower risk group was longer, with 6.93 years (5.56−not available [NA]) compared with the higher risk group, with 3.26 years (2.68−3.81) years. Thus, the latter group had significantly inferior OS (p = 4.38 × 10−6) and higher death rate (p = 3.08 × 10−4). The high-risk group was enriched with patients showing the following characteristics:
- SMF
- JAK2V617F homozygous mutations
- ≥1 HMR mutation
- Older median age
- Elevated >1% or 2% circulating blasts
- Splenomegaly
- Bone marrow fibrosis Grade ≥2
Gene expression profile-based model
Based on low- and high-risk categories from hierarchical clustering, 201 genes were identified for the GEP based model. Patients were, again, classified into two risk groups:
- Survival rate of high-risk group was significantly lower, i.e., 3.26 years (2.68−3.81) compared with 6.93 years (5.56−NA) in the low-risk cohort (p = 1.78 × 10−7).
- Median age at the time of sample collection for high-risk group was higher (69.95 years) compared with low-risk group (61.40 years; p = 5.73 × 10−5).
- High-risk group was enriched in patients with PET-MF and PPV-MF while patients with pre-PMF dominated low-risk group.
- Overt PMF cases were similar between the two risk groups.
- The presence of the homozygous JAK2V617F mutation, ASXL1 mutation, and ≥1 HMR mutations were greater in the high-risk group.
It was observed that high-risk classification was linked to the presence of clinical markers of inferior survival. Higher patient age at the time of sample collection, lower hemoglobin (11.15 g/dL vs 12.1 in low-risk group, p = 3.98 × 10-6) and platelet counts, higher white blood cell (WBC) counts (14.2 × 109/L vs 8.0 × 109/L in low-risk group; p = 9.44 × 10-3) and increased incidence of splenomegaly, presence of circulating blasts ≥1% or ≥2%, bone marrow fibrosis Grade ≥2, and constitutional symptoms were observed in this group (Figure 1). Furthermore, leukemia-free survival in the high-risk group was significantly low (p = 1.9 × 10−2).
High-risk profile was associated with a risk for lower survival (HR, 4.736; 95% confidence interval [CI], 2.5−8.9; p = 1.48 × 10−6) and leukemic transformation (HR, 3.976; 95% CI, 1.2−13.6; p = 2.75 × 10−2).
Figure 1. Clinical and molecular characteristics based on gene expression.*
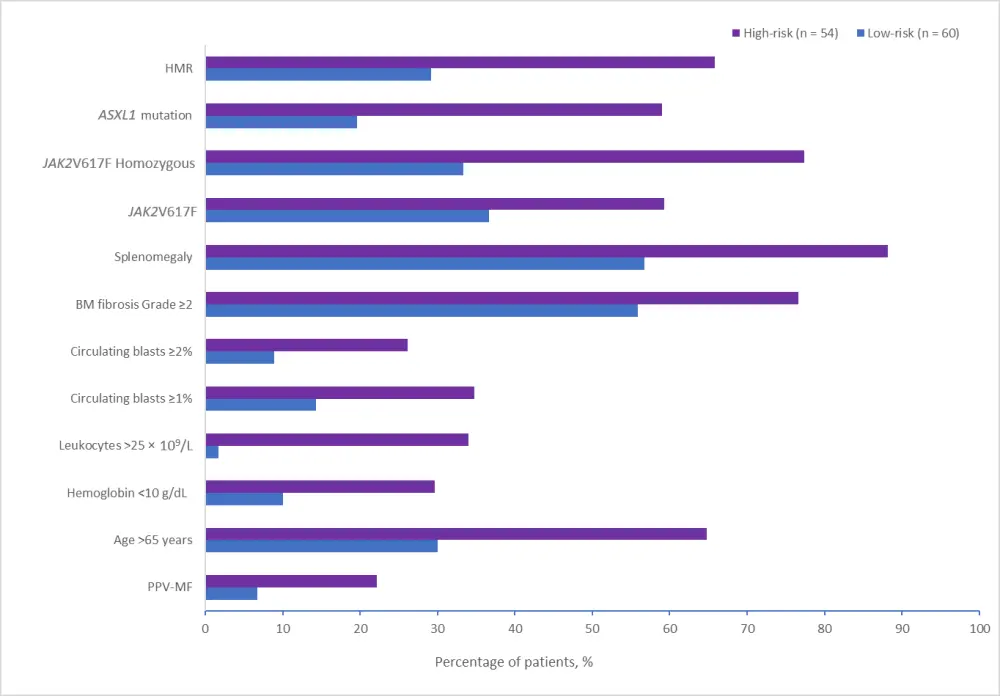
ASXL1, polycomb chromatin-binding protein gene; BM, bone marrow; HMR, high molecular risk; JAK2V617F, Janus kinase 2 mutation in phenylalanine at position 617; PPV-MF, post-polycythemia vera myelofibrosis.
*Adapted from Rontauroli et al.1
GEP-based model vs contemporary prognostic model
- Patient distribution for GEP based model was different from DIPSS and MIPSS70.
- Patients from low- and intermediate-1 category based on the DIPSS model were classified as low-risk, whereas DIPSS intermediate-2, and high-risk patients were grouped in the high-risk in the GEP model.
- In terms of MIPSS70, the high-risk group retained the same high-risk status in GEP, while most patients from intermediate- and low-risk groups were designated as low-risk in the GEP model.
- Interestingly, the GEP high-risk classification remained a significant independent variable in multivariate analysis for the DIPSS and MIPSS70 classifications (p = 4.96 × 10−3 and 1.12 × 10−4, respectively), and risk factors included in the DIPSS and MIPSS70 models (p = 6 × 10−4 and 1.01 × 10−4, respectively) (Table 2).
Table 2. Multivariate regression analysis of prognostic factors for OS in patients classified according to DIPSS/MIPSS70 model*
|
DIPSS, Dynamic International Prognostic Scoring System; MIPSS70, mutation-enhanced International Prognostic Scoring System; OS, overall survival. |
|||
|
Variable |
Low-risk |
High-risk |
p value |
|---|---|---|---|
|
DIPSS (n evaluable, total = 107) |
53 |
47 |
— |
|
Low |
40.4 |
14 |
— |
|
Intermediate-1 |
42.1 |
34 |
— |
|
Intermediate-2 |
14 |
32 |
— |
|
High |
3.5 |
20 |
6.00 × 10−4 |
|
MIPSS70 (n evaluable, total = 73) |
56 |
44 |
— |
|
Low |
29.3 |
3.1 |
— |
|
Intermediate |
53.7 |
34.4 |
— |
|
High |
17.1 |
62.5 |
1.01 × 10−4 |
- The new model allowed to identify GEP high- and low-risk patients in the DIPSS intermediate-1 and intermediate-2 groups, and to better predict their survival. Similarly, it distinguished GEP high-risk patients from those with low-risk, predicting different survival within MIPSS70 intermediate- and high-risk categories (Table 2).
Conclusion
The GEP-based risk classification used in this study was identified as an independent predictive factor for survival, which allowed to further distinguish patients with lower survival within intermediate risk classes on contemporary risk classification systems. The study also validates the concept that genetic expression is associated with molecular and clinical characteristics. Therefore, genetic expression profiling can be used to complement the traditional risk stratification tools and be helpful to predict the survival outcomes of MF patients.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

