All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MPN Advocates Network.
The mpn Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mpn Hub cannot guarantee the accuracy of translated content. The mpn and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The MPN Hub is an independent medical education platform, sponsored by AOP Health, GSK, Sumitomo Pharma, and supported through educational grants from Bristol Myers Squibb and Incyte. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MPN content recommended for you
Gene panel testing for MDS/MPN overlap
A diagnosis of myelodysplastic syndromes/myeloproliferative neoplasms (MDS/MPN) overlap is given when a patient exhibits morphological features of both dysplasia and myeloid proliferation. However, precision in diagnosis is difficult owing to the heterogeneity of these clonal hematopoiesis disorders. As such, the identification of chromosomal and clonal abnormalities may help to differentiate diagnosis and improve treatment individualization.
In a recent review by Anoop K. Enjeti et al.,1 published in Pathology, experts in MDS and MPN provided a recommended gene panel to be integrated into the diagnosis and prognosis of MDS/MPN clonal disorders. We summarize the key guidance below.
Recommended approach to using gene panel testing for MDS/MPN
In adults, there are five main subtypes of MPN/MDS overlap: atypical chronic myeloid leukemia (aCML); chronic myelomonocytic leukemia (CMML); MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T); MDS/MPN-unclassifiable (MDS/MPN-U); and therapy-related MDS/MPN (t-MDS/MPN). Incorporation of genetic testing in these disorders is used for risk stratification/prognostic scoring systems, and the most relevant mutations are shown in Table 1.
Table 1. Somatic mutations relevant to diagnosis and prognosis of MPN/MDS overlap*
|
aCML, atypical chronic myeloid leukemia; CMML, chronic myelomonocytic leukemia; CPSS-Mol, CMML Prognostic Scoring System-Molecular; MDS, myelodysplastic syndromes; MPN, myeloproliferative neoplasms; MDS/MPN-RS-T, MDS/MPN with ring sideroblasts and thrombocytosis; MDS/MPN-U, MDS/MPN-unclassifiable. |
||
|
Overlap syndrome |
Somatic mutation |
Somatic mutations with prognostic relevance or included in risk stratification/prognostic scoring systems |
|---|---|---|
|
CMML |
TET2 (~60%) SRSF2 (~50%) ASXL1 (~40%) SETBP1 Concurrent TET2/SRSF2/RUNX1 (10–20%) TP53 (~1%) |
Mayo molecular model (ASXL1 frameshift or nonsense mutation) CPSS-Mol (ASXL1, NRAS or SETBP1 and RUNX1 mutations) Groupe Francophone des Myélodysplasies (ASXL1 mutation) |
|
aCML |
ASXL1 (60%) SETBP1 (~20–40%) ETNK1 (~8%) NRAS/KRAS (35%) |
Mayo prognostic model for aCML (TET2 mutation) SETBP1 |
|
MDS/MPN-RS-T |
SF3B1 (~90%) JAK2 V617F (50%) CALR (<5%) MPL (<5%) DNMT3A (~10–30%) SETBP1 (~10%) |
Mayo prognostic model for MDS/MPN-RS-T (ASXL1 or SETBP1 mutations) |
|
MDS/MPN-U |
ASXL1 (~30–55%) TET2/SRSF2/JAK2 (~25%) SETBP1/EZH2/NRAS/KRAS (~10–15%) |
ASXL1 is associated with poor prognosis Overall prognosis is more dependent on cytogenetic and molecular features of individual cases |
Certain mutational combinations are more common with certain subtypes of MDS/MPN, and when clonal features are combined with morphological identifiers, a more accurate picture of the subtype and disease course may be captured. The expert panel, led by chairs of the MDS, MPN, and working parties of the Australasian Leukaemia & Lymphoma Group (ALLG), provided recommended guidance for diagnosis and prognosis of MDS/MPN. Notably, gene testing does not drive differentiation, but rather acts as a supportive role in addition to these features for both diagnosis and prognosis (Figure 1). The authors also highlighted the possibility of using machine learning approaches when combining genotype-phenotype associations.
Figure 1. Recommended guidance for diagnosed MDS/MPN subtypes*†
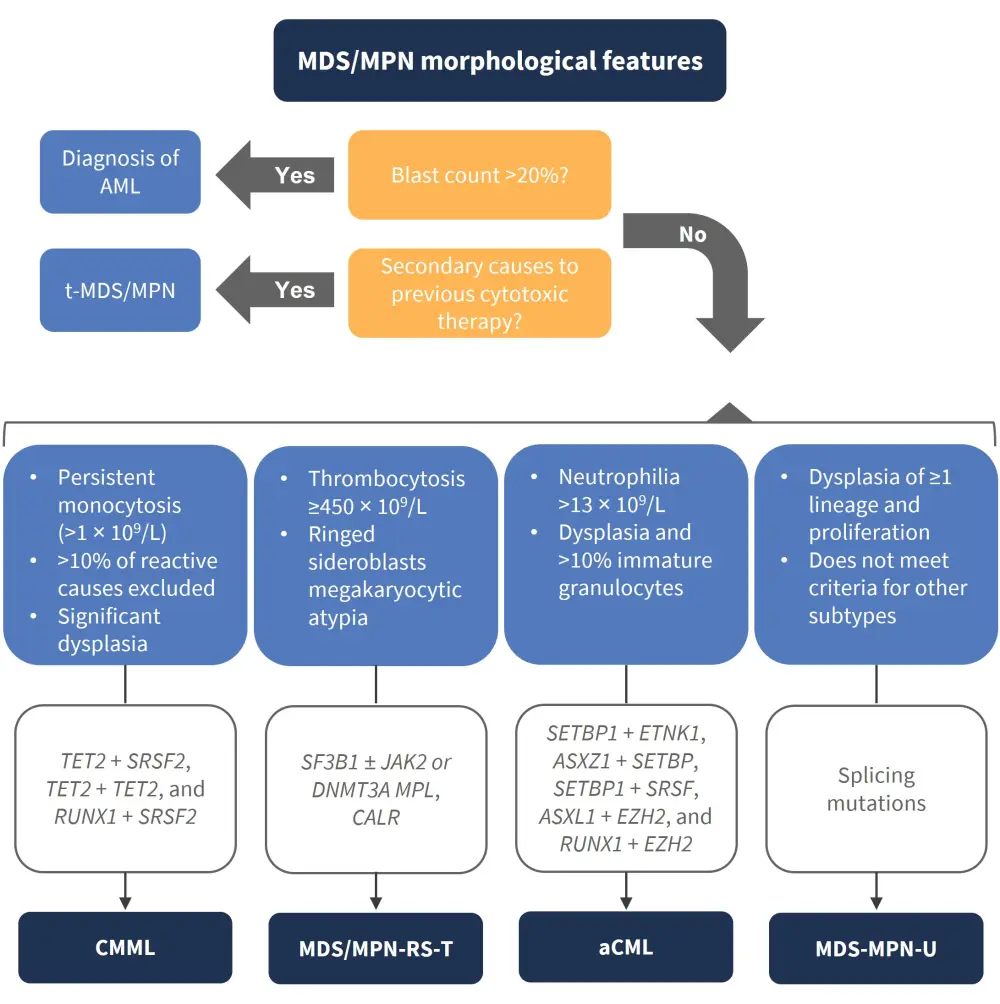
aCML, atypical chronic myeloid leukemia; AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia; MDS, myelodysplastic syndromes; MDS/MPN-R-T, MDS/MPN with ring sideroblasts and thrombocytosis; MDS/MPN-U, MDS/MPN-unclassifiable; MPN, myeloproliferative neoplasms; T-MDS/MPN, therapy-related MDS/MPN.
*Adapted from Enjeti et al.1
†Consensus panel of genes considered supportive in diagnosis, alongside morphological features of MDS/MPN: SF3B1, TET2, SRSF2, ASXL1, JAK2, NRAS, KRAS, CBL, RUNX1, SETBP1, MPL, CALR, ETNK1, DNMT3A, IDH1‡, IDH2; and genes with prognostic relevance: TP53, EZH2, U2AF1.
‡Rare in MDS/MPN.
Conclusion
The recommended gene panel for integration in the diagnosis and prognosis of MDS/MPN demonstrates the substantial heterogeneity of these disorders. Using this large data analysis approach, more individualized therapeutic guidance may be possible in MDS/MPN.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

