All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MPN Advocates Network.
The mpn Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mpn Hub cannot guarantee the accuracy of translated content. The mpn and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The MPN Hub is an independent medical education platform, sponsored by AOP Health, GSK, Sumitomo Pharma, and supported through educational grants from Bristol Myers Squibb and Incyte. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MPN content recommended for you
Single-cell analysis at ASH: Drivers of leukemic transformation
Around 10–20% of patients with myeloproliferative neoplasms (MPN) eventually develop secondary acute myeloid leukemia (sAML) and are faced with a poor prognosis. Certain mutations involved in leukemic evolution have been identified, but single-cell analysis promises to unpick clonal complexity and provides details of the heterogenous cell populations at different disease timepoints.1
At the 63rd American Society of Hematology (ASH) Annual Meeting and Exposition, two presentations detailed the findings from single-cell studies on leukemic transformation.1,2
Leukemic transformation in TP53-mutated MPN2
Mutations in TP53 are observed in a proportion of chronic phase MPN. Interestingly, of the cases that acquire TP53 mutations, there is around a 50/50 split in those that progress to aggressive sAML and those that remain in chronic phase MPN. This characteristic provides a robust cell model to investigate the drivers of leukemic transformation in TP53-mutated cells.
Application of single-cell multiomics allows detailed analysis of heterogenous tumor microenvironments, such as those observed in acute phase hematologic malignancies. Alba Rodriguez-Meira, University of Oxford, UK, presented data from a preclinical study evaluating tumor evolution, cellular hierarchies, and molecular features of TP53-driven transformation by single-cell analysis.2
Study design
The study processed 22,116 cells from 35 donors (Figure 1) at 40 different disease timepoints. Lin−CD34+ cells were isolated and evaluated using TARGET-seq analysis to detect mutations, cell surface proteins, and transcriptomic information from individual cells. A threshold of >95% allelic resolution was used to investigate different TP53 zygosity status and identify wildtype (WT) cells.
Figure 1. Study donors*
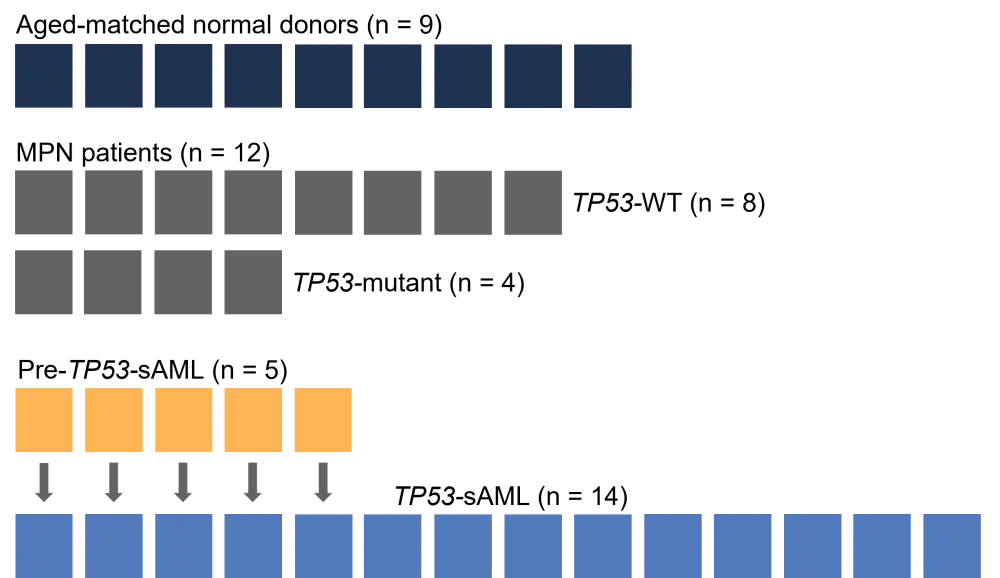
AML, acute myeloid leukemia; MPN, myeloproliferative neoplasms; sAML, secondary AML; WT, wildtype.
*Adapted from Rodriguez-Meira et al.2
Leukemic transformation: Genetic clonal evolution
As the first objective, Rodriguez-Meira and colleagues2 set out to identify the clonal hierarchies associated with acute disease. The team uncovered the initial MPN driver or associated mutations, followed by loss of TP53 WT and subsequent acquisition of multi-hit TP53 mutations throughout the course of transformation. The latter was observed in 100% of patients and three key characteristics of clonal evolution were established: biallelic TP53 mutations (n = 6), hemizygous TP53 mutations (n = 6), and simultaneous biallelic and hemizygous TP53 mutations (n = 2). Further analysis of the TP53 mutated multi-hit cell population highlighted the presence of copy number abnormalities in >80% of cells in all of the patients evaluated. This analysis also uncovered the presence of small cytogenetic subclones that were able to multiply in patient-derived xenograft mouse models. Therefore, when occurring together, mutations in p53, loss of TP53 WT, and complex cytogenetic evolution represent the driving factors of leukemic stem cell transformation.
Leukemic transformation: Molecular hierarchies
Transcriptomic analysis identified the main cell types within the sAML population:
- TP53-mutant erythroid biased cells
- TP53-mutant leukemic stem cells (LSCs)
- Pre-leukemic TP53-WT cells
An erythroid-biased cell cluster was considered an unexpected observation as these patients were not diagnosed with erythroid leukemia. This observation was further validated by using a normal donor hematopoietic atlas; TP53-mutant sAML cells showed clustering in hematopoietic stem cells and early erythroid progenitors. In addition, the erythroid score was significantly higher in mutant cells versus WT cells.
The TP53-mutant LSC cluster exhibited a mutational pattern that provided the basis for a new prognostic tool. In sAML and de novo AML (BeatAML cohort), the LSC score was prognostic regardless of TP53 mutation status, highlighting the value of single-cell multiomics in differentiating cell populations in acute disease and providing clinical and prognostic insights.
Pre-leukemic TP53-WT cells underwent further evaluation against cells from normal donors and patients with myelofibrosis. TARGET-seq analysis uncovered elevated stem-cell associated transcription and self-renewal, quiescence, and low differentiation capacity, suggesting cell-intrinsic suppression.
Leukemic progression
Finally, the study compared the molecular characteristics of patients with TP53-mutant MPN who did/did not progress to sAML. The data suggest comparable levels of TP53 heterozygous clones between the two populations (Figure 2), but transcriptomic analysis highlighted an upregulation of interferon response genes and inflammatory signaling in patients who progress to acute leukemia. In vivo mouse models were used to confirm the role of chronic inflammation in TP53-mutant clonal expansion and progression to acute leukemia.
Figure 2. Clonal characteristics of disease progression from MPN to acute myeloid leukemia*
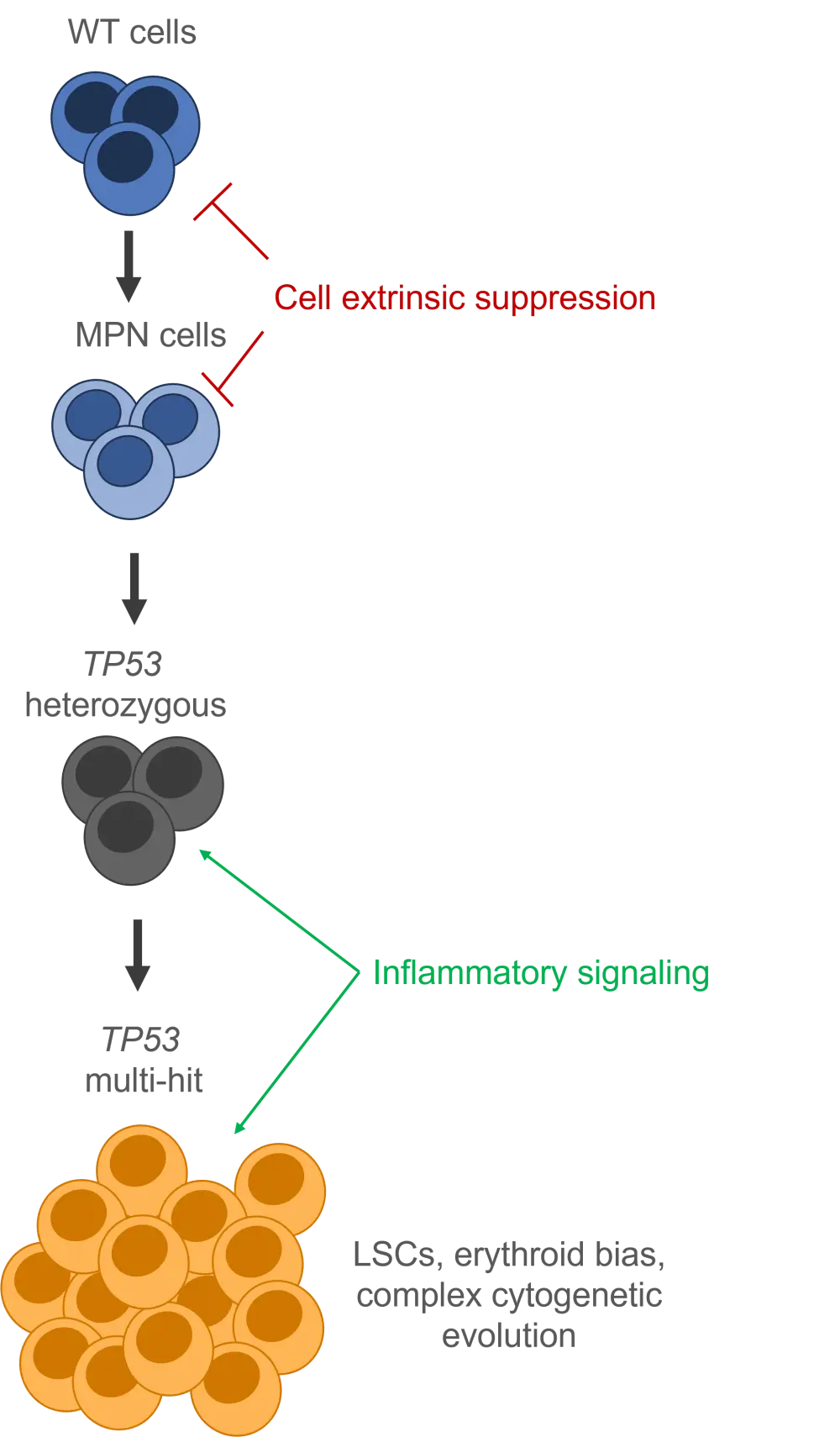
AML, acute myeloid leukemia; MPN, myeloproliferative neoplasms; WT, wildtype.
*Adapted from Rodriguez-Meira et al.2
Taken together, data from this study has provided a model of progression from MPN to aggressive leukemia (Figure 3).
Figure 3. Proposed model for leukemic progression from MPN to sAML*
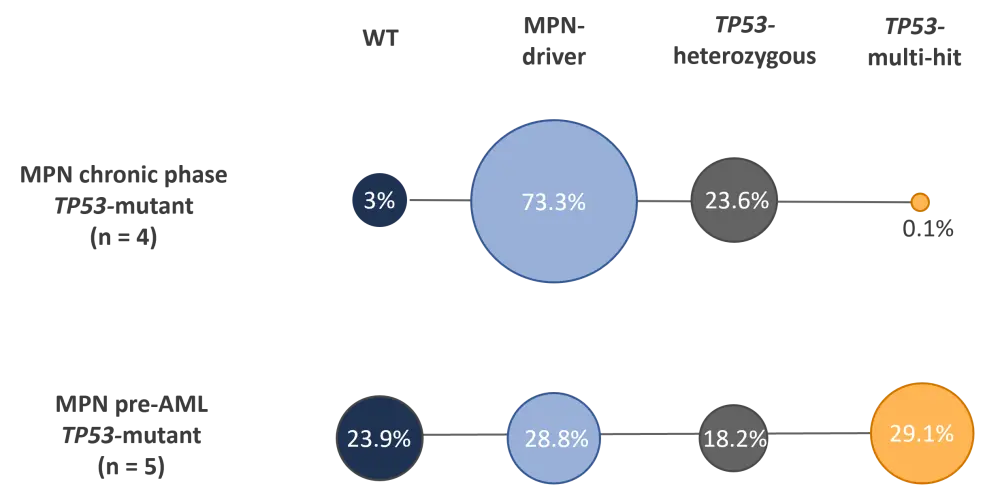
LSCs, leukemic stem cells; MPN, myeloproliferative neoplasms; sAML, secondary acute myeloid leukemia; WT, wildtype.
*Adapted from Rodriguez-Meira et al.2
Mutational evolution of MPN in leukemic progression1
In a separate study, Paola Guglielmelli et al.1 sought to identify mutation patterns of leukemic transformation and determine markers unique to blast-phase MPN as it shows distinct features than de novo AML.
Study design
A total of 22 paired samples were obtained from 10 patients with MPN in chronic and blast phases. The CD34+ cells underwent next-generation sequencing (NGS) target resequencing and single-cell multiomic genotyping.
Bulk analysis: Chronic vs blast phase
Bulk analysis of chronic phase and blast phase MPN samples identified mutations in both coding somatic regions and epigenetic regulators, and additional mutations were identified in samples from blast phase disease (Figure 4).
- The numbers of coding somatic mutations were 26 and 34 in chronic and blast phase, respectively.
- There were 16 (61.5%) and 23 (67.6%) single-nucleotide variants detected in chronic and blast phase, respectively.
Figure 4. Mutations identified in MPN cells in A chronic and B blast phase disease by bulk analysis*
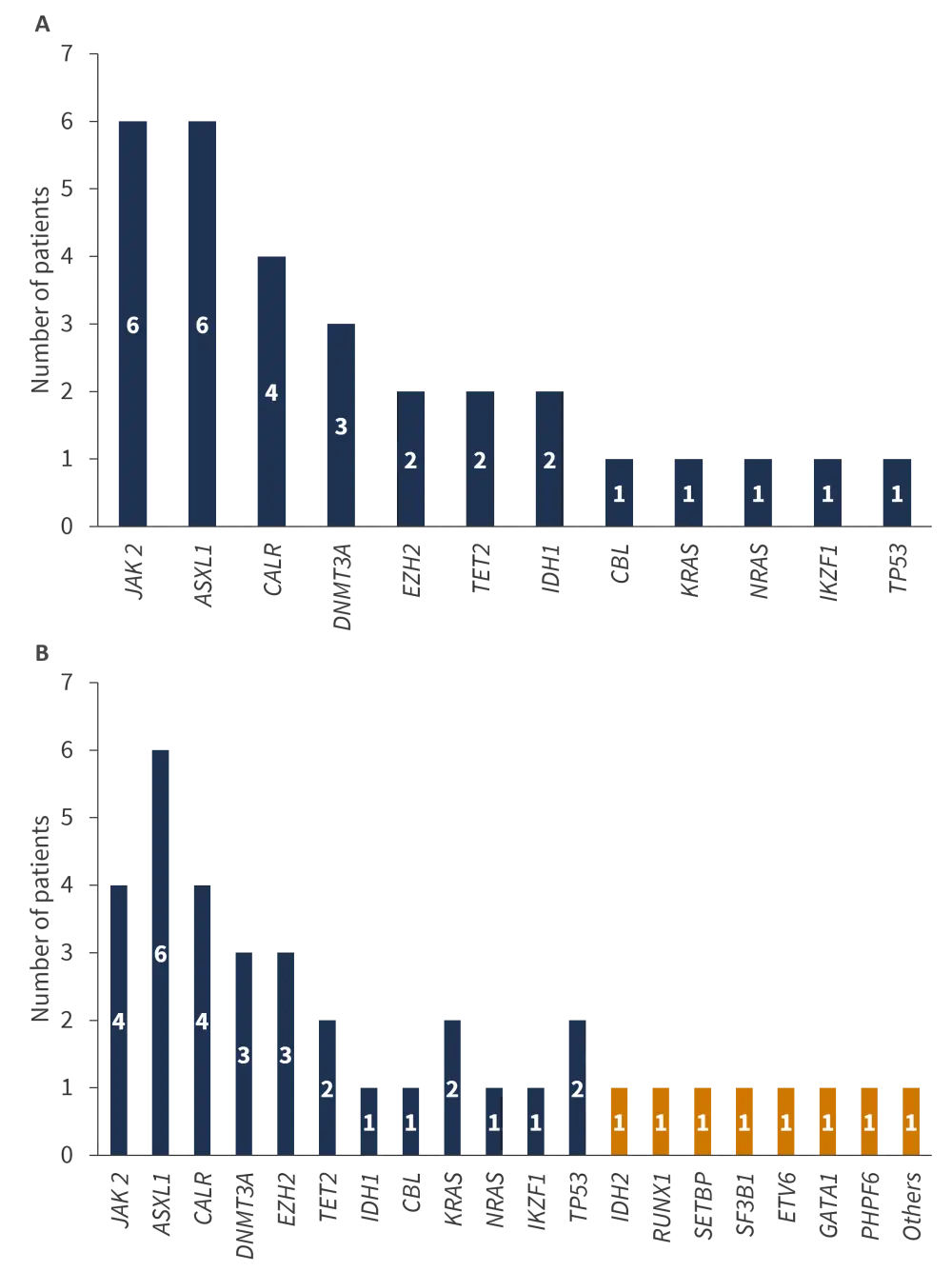
MPN, myeloproliferative neoplasms.
Orange bars represent additional mutations observed in blast phase disease.
*Adapted from Guglielmelli et al.1
Single-cell analysis: Chronic vs blast phase
Single-cell analysis was used to further investigate the clonal landscape at the different disease stages. The number of mutated clones increased from 2–3 in chronic phase to 2–5 in blast phase disease. Single-cell analysis was successful at identifying dominant leukemic clones in 7 of the 10 patients, a finding overlooked by bulk sequencing, and was also more sensitive to clonal heterogeneity, with 36% and 17% further variants detected in chronic and blast phase, respectively. Importantly, single-cell analysis identified significant differences in copy number variations between chronic and blast-phase samples. Regional amplification of a number of genes were associated with blast phase disease, including ETV6 in 6 patients (≥20 copies in 2 patients), RUNX1 in 3 patients (≥6 copies), and BRAF in 4 patients (≥3 copies). Additionally, gain in MPL, and loss of EZH2 and ZHSR2 were characteristic of cells in blast phase.
Conclusion
Across the two studies, single-cell analysis has assisted in defining the clonal evolution of leukemic transformation from MPN to sAML. Moving forward, single-cell analysis may assist in the identification of therapeutic targets to prevent the transformation of MPN to aggressive disease, and eventually influence treatment decisions.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

