All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MPN Advocates Network.
The mpn Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mpn Hub cannot guarantee the accuracy of translated content. The mpn and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The MPN Hub is an independent medical education platform, sponsored by AOP Health, GSK, Sumitomo Pharma, and supported through educational grants from Bristol Myers Squibb and Incyte. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MPN content recommended for you
Whole-genome sequencing shows driver mutations can be acquired very early in life
During the 62nd American Society of Hematology (ASH) Annual Meeting and Exposition, Jyoti Nangalia presented intriguing findings in adult patients with myeloproliferative neoplasms (MPN) based on a study conducted by Nicholas Williams and colleagues that shed light on the timing of driver mutation acquisition and clonal dynamics. Here, we are pleased to give an overview on this study and summarize exciting results.
For capturing early cancer development, the authors took advantage of the unique nature of MPN where normal hematopoiesis and cancer clones coexist in the bone marrow. In addition, they were helped by the well-characterized genetic mutational landscape in MPN where most patients have been shown to harbor a driver mutation in the genes of either JAK2, calreticulin (CALR), or thrombopoietin receptor (MPL), and more than half have mutations in additional cancer-related genes, such as TET2, DNMT3A, or ASXL1.
Study design
A total of ten patients aged between 20 and 76 years with different subtypes, i.e., essential thrombocythemia (ET), polycythemia vera (PV), and post-PV myelofibrosis, were included in the study. Samples were collected from either peripheral blood or bone marrow tissue to obtain single-cell derived hematopoietic colonies. Each colony underwent whole-genome sequencing (n = 952), which reflected the single-cell from which the colony was derived.
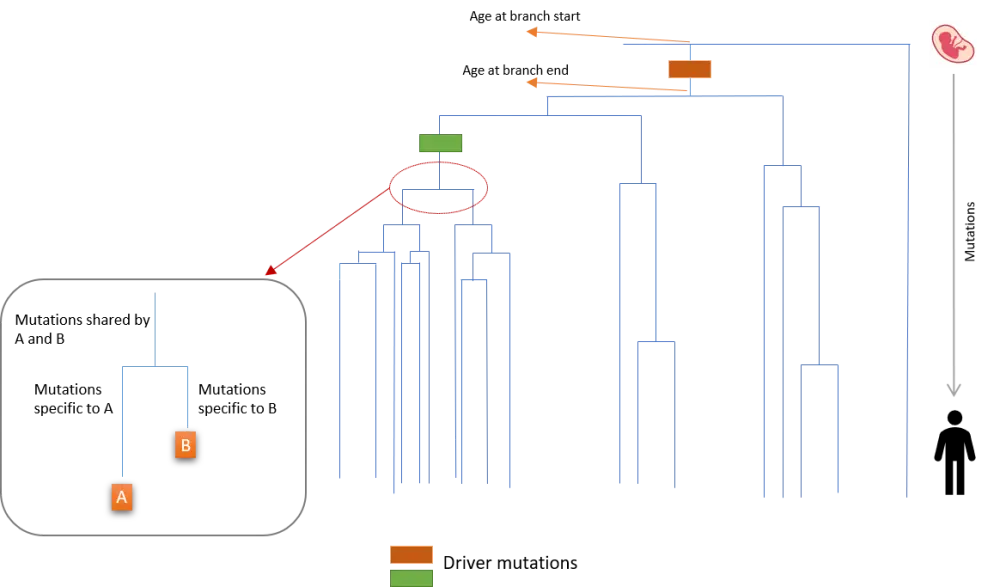
Whole-genome sequencing at the clonal level demonstrated that there was an abundance of MPN- and cancer-related driver mutations, as well as a higher-than-expected level of copy number changes compared with levels from bulk whole exome sequencing. Pattern of sharedness or absence across colonies was analyzed using a phylogenetic tree where somatic mutations (n = 448,553) and driver mutations were placed on the branches according to their presence in each colony (see Figure 1). This provided information on the sequence of driver mutation appearance in each patient.
Furthermore, as blood stem cells acquire about 18 somatic mutations per year across the genome, the authors were able to convert the relative timing of driver mutation acquisition to absolute timing, correlating somatic mutations in each colony with patient’s age (Figure 1). The pattern of branching demonstrated the expansion of mutated stem cells into clones, and the repetitive branching showed how fast a mutated clone was expanding over time.
Figure 1. Branches on the phylogenetic tree (adapted from Williams et al.1)
Results
Detecting mutated clones
The analysis of the phylogenetic tree of a patient who was diagnosed with ET at the age of 21 years and sampled at 23 years showed that mutant clones had a common ancestor harboring a JAK2 V617F mutation, and shared about 80 mutations, while wild-type hematopoietic cells showed only minimal sharing of mutations. The estimated age at the start and end of the JAK2 mutation acquisition corresponded to 6.2 weeks post-conception and 1.3 years of age, respectively, indicating that the early acquisition of a driver mutation happened decades before diagnosis.
Whole-genome sequencing of another patient who was diagnosed with PV at 31 years and sampled at 38 showed that the JAK2 V617F mutation was acquired during early childhood (between 4.2 weeks post-conception and 8.6 years of age) inducing a rapidly expanding clone. Interestingly, there was also a DNMT3A mutation acquired within gestation that expanded only very slowly leading to a very small clonal fraction in later life. Similarly, another patient’s DNMT3A mutations could be dated to the early weeks post gestation. DNMT3A mutations belong to the most common mutations found in acute myeloid leukemia and clonal hematopoiesis.
Some patients developed a JAK2 V617F mutation as the second driver mutation. Two patients who were diagnosed with ET were shown to have acquired the DNMTA3 mutations first, followed by a JAK2 mutation many years later, and it took many years to finally develop the disease. Interestingly, the oldest patient in the study population, with an age of 76 years, had a driverless clonal expansion that started many years before diagnosis.
One patient who was diagnosed with PV at 53 years and sampled at 68 years, progressed to MF at the age of 71 years. Clonal analysis showed that one JAK2 mutation was acquired in childhood, while three independent DNMTA3 mutations, and four independent homozygous JAK2 mutations occurred later in life. These findings indicated that disease progression can evolve through multiple independent paths, and that other factors such as germline background or bone marrow microenvironment may predispose patients for certain driver mutations.
Measuring growth rate of mutated clones
To evaluate the expansion rate of the clones, mutations identified in the first part of the study were re-sequenced in multiple longitudinal blood samples. Then branching pattern and mutation fractions from blood samples were used for a combined analysis.
Results showed that growth rates of clones with driver mutations varied significantly, suggesting that additional mutations influenced growth rates. In one patient with a DNMT3A mutation, the addition of other mutations increased cell growth rate per year as follows:
- 9% with DNMT3A alone
- 67% with JAK2 V617F and 9pUPD
- 233% with JAK2 V617F, 9pUPD, and TET2
When individual patients with JAK2 V617F mutations were compared, the differences in growth rate per year varied between 18% and 68%, indicating that patient-related factors impacted on growth rates.
As was to be expected, a slower growth rate delayed diagnosis. The mean latency between JAK2 V617F mutation and disease presentation was 34 years (range, 20–54 years).
Conclusion
This study showed that JAK2 and DNMTA3 mutations can be acquired very early in life, starting already in utero for DNMTA3 mutation. The growth rate varied among different clones, and patient-specific factors such as mutations contributed to these differences. The fact that slower growing mutations were associated with a longer latency to diagnosis indicates that individuals with mutant clones need to be surveyed over time to assess clonal expansion rate and that intervention should happen early if expansion accelerates.
The authors concluded that early detection of mutation could detect predisposition for a disease, which would ultimately inform disease prevention strategies. Further evaluation is needed in a larger sample size to confirm these findings and to assess their application in other areas, such as pediatric cancers.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

