All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MPN Advocates Network.
The mpn Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mpn Hub cannot guarantee the accuracy of translated content. The mpn and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The MPN Hub is an independent medical education platform, sponsored by AOP Health, GSK, Sumitomo Pharma, and supported through educational grants from Bristol Myers Squibb and Incyte. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MPN content recommended for you
Applying prognostic models for myelofibrosis in practice
The overall survival (OS) of patients with primary myelofibrosis (PMF) varies widely, depending on individual clinical and genetic factors. The most common causes of death in PMF are hematologic transformation, disease progression, and infections, along with other causes of morbidity and mortality. Given the heterogeneity of the disease, precise PMF models are essential for individual estimation of disease outcome and management. Consequently, there has been considerable effort to develop discriminatory prognostic models to help clinical decision making in several settings, including hematopoietic stem cell transplantation (HSCT).
The review by How et al.1 covers established and emerging prognostic models for PMF, such as the International Prognostic Scoring System (IPSS); Dynamic IPSS (DIPSS) and its revision DIPSS+; the Mutation-Enhanced IPSS for transplant-age patients (MIPSS70) and MIPSS70+ (containing cytogenetic information); the Genetically-Inspired Prognostic Scoring System (GIPSS); and the Myelofibrosis Secondary to PV and ET-Prognostic Model (MYSEC-PM) for patients with myelofibrosis occurring post-essential thrombocytosis (ET) or polycythemia vera (PV). Moreover, the authors provide guidance on how to use these prognostic models in clinical practice.
Contemporary models1
International Prognostic Scoring System
The IPSS was established based on data from 1,054 patients with PMF to help with prognostication and treatment decisions after diagnosis. The IPSS comprises of five variables: age > 65 years, hemoglobin (Hb) level < 10 g/dL, white blood cell count > 25 GPT/L, circulating blasts ≥ 1%, and presence of constitutional symptoms. Depending on the number of variables present, patients are subdivided into low-, intermediate-1, intermediate-2, and high-risk groups, corresponding with decreasing median OS. Of note, cytogenetic variables are not included.2
Dynamic IPSS and Dynamic IPSS+
In contrast to the IPSS, the DIPSS is a PMF model that can be used at any time of disease assessment. Using a study analyzing 525 patients with PMF, the DIPSS validated the impact of various risk factors on OS: for example, as anemia had the most substantial impact on OS, it was assigned two points within this system. Subsequent research showed that the DIPSS also had predictive value for leukemic transformation. When further prognostic factors were identified, such as red blood cell transfusion, unfavorable karyotype, and thrombocytopenia, they were included in DIPPS+. For assessing leukemia-free survival (LFS), only thrombocytopenia and unfavorable karyotype were identified as significant factors.1-4 The DIPPS and DIPPS+ remain the cornerstones of PMF prognostications due to their easy use and basis on readily available clinical data.
Mutation-Enhanced IPSS
Numerous mutations in myeloproliferative neoplasms (MPN) have been identified in recent years. The inclusion of these high-risk mutations led to the development of multiple genetic-based scoring systems, such as MIPSS70, without cytogenetic information, the MIPSS70+ with cytogenetic information included, and MIPSS70+ version 2.0 (v2.0).
The MIPSS70 includes both clinical and molecular risk factors (Table 1). As the MIPSS70 was specifically designed for the transplant setting, it is therefore age restricted. The MIPSS70 model groups patients into low-, intermediate-, or high-risk categories with a corresponding median OS of 27.7, 7.1, and 2.3 years, respectively. It is also predictive for LFS. The MIPSS70+ incorporates three clinical factors, considers high-risk mutations and karyotype, and places more weight on unfavorable cytogenetic information (Table 1). Recently, MIPSS70+ was updated to MIPSS70 v2.0 with newer genetic findings and differentiation of patients with an unfavorable versus a very high-risk karyotype, adding U2AF1Q157 as a high molecular risk (HMR) mutation (Table 1).
Table 1. Prognostic models for PMF and post-ET/PV MF1
|
Prognostic model |
Variables (points awarded) |
Risk groups (median OS, years) |
Unfavorable karyotype |
VHR karyotype |
|---|---|---|---|---|
|
BM, bone marrow; DIPSS, dynamic IPSS; ET, essential thrombocythemia; GIPSS, genetically-inspired prognostic scoring system; Hb, hemoglobin; HLA, human leukocyte antigen; HMR, high molecular risk; int, intermediate; IPSS, International Prognostic Scoring System; KPS, Karnofsky performance status; MF, myelofibrosis; MIPSS70, mutation-enhanced prognostic scoring system for transplant-age patients; MIPSS70+ v2.0, MIPSS70 plus version 2.0; MPN, myeloproliferative neoplasms; MTSS, Myelofibrosis Transplant Scoring System; MYSEC-PM, Myelofibrosis Secondary to PV and ET-Prognostic Model; OS, overall survival; PMF, primary myelofibrosis; PV, polycythemia vera; VHR, very high risk; WBC, white blood cell. |
||||
|
IPSS2 |
Age > 65 (1) |
0 = low (11.3) |
Not scored |
Not scored |
|
DIPSS3 |
Age > 65 a (1) |
0 = low (not reached) |
Presence of complex karyotype or sole or 2 abnormalities that included 18, 27/7q2, i(17q), inv(3), 25/5q2, 12p2, or 11q23 |
Not scored |
|
DIPSS+4 |
DIPSS low (0) |
0 = low (15.4) |
Not scored |
Not scored |
|
MIPSS705 |
Hb < 10g/dL (1) |
0–1 = low (27.7) |
Not scored |
Not scored |
|
MIPSS70+5 |
Hb < 10 g/dL (1) |
0–2 = low (20) |
Any karyotype other than normal karyotype or sole abnormalities of 20q-, 13q-, 19, chromosome 1 translocation/duplication, -Y, or sex chromosome abnormality other than -Y |
Not scored |
|
GIPSS7 |
VHR karyotype (2) |
0 = low (26.4) |
Any karyotype other than very high-risk karyotype, normal karyotype, or sole abnormalities of 20q-, 13q-, 19, chromosome 1 translocation/duplication, -Y, or sex chromosome abnormality other than -Y |
Single or multiple abnormalities of -7, i(17q), inv(3)/3q21, 12p-/12p11.2, 11q-/11q23, or other autosomal trisomies not including 18/19 |
|
MIPSS70+ v2.06 |
VHR karyotype (4) |
0 = very low (26.4) |
Any karyotype other than very high-risk karyotype, normal karyotype, or sole abnormalities of 20q-, 13q-, 19, chromosome 1 translocation/duplication, -Y, or sex chromosome abnormality other than -Y |
Single or multiple abnormalities of -7, i(17q), inv(3)/3q21, 12p-/12p11.2, 11q-/11q23, or other autosomal trisomies not including 18/19 |
|
MYSEC-PM8 |
Age (0.15 per year of age) |
< 11 = low (not reached) |
Not scored |
Not scored |
|
MPN Personalized Risk Calculator9 |
For variables and scoring, see link |
Individualized risk groups |
— |
— |
|
MTSS10 |
Age ≥ 57 years (1) |
0–2 = low (5-year OS, 90%) |
Not scored |
Not scored |
Genetically-Inspired Prognostic Scoring System
The GIPSS reflects a purely genetic prognostic model and was developed to complement the MIPSS70+ v2.0. The GIPSS categorized patient karyotype as very-high-risk, unfavorable, or favorable (Table 1). The GIPSS was non-inferior to the MIPSS70+ and DIPSS and was validated for OS and LFS in a reference cohort.
Newly emerging models1
Recent work analyzed 69 cancer genes in 2,000 patients with MPN by whole-genome sequencing. Interestingly, p53 mutations were associated with an inferior outcome. The model also integrated clinical and cytogenetic variables to create a more individualized prediction of OS and risk for transformation to acute myeloid leukemia. This MPN Personalized Risk Calculator provides a patient-specific approach based on cancer genomics.
The Myelofibrosis Transplant Scoring System (MTSS) was established to predict the prognosis for MPN patients potentially undergoing HSCT. The MTSS was evaluated in patients with PMF and post-ET/PV MF who underwent HSCT and found unfavorable outcome variables, such as age ≥ 57 years, Karnofsky performance status < 90%, platelet count < 150 Gpt/L, white blood cell count > 25 Gpt/L, HLA-mismatched donor, ASXL1 mutation, and non-CALR/MPL driver mutation. Other variables in a pre-transplant setting (constitutional symptoms, cytogenetic risk, Hb level, ≥ HMR mutations) were not found to be prognostic. The MTSS was more accurate in predicting outcomes in HSCT patients than DIPSS, MIPSS70, and MYSEC-PM.
Application of prognostic models in clinical settings1
Currently, guidelines recommend the use of IPSS, DIPSS, and DIPSS+ for risk stratification in PMF. Although these models are easy to apply in daily clinical practice, risk stratification approaches that incorporate molecular and cytogenetic information such as MIPSS70+ v2.0 provide better risk discrimination, especially for patients with intermediate-2 to high-risk classification according to IPSS, DIPSS, and DIPSS+ who are candidates for HSCT. Also, scoring systems that consider genetic and cytogenetic data seem to have increased prognostic value to estimate the risk of leukemia progression (Figure 1).
Figure 1. Risk stratification approach in primary myelofibrosis1
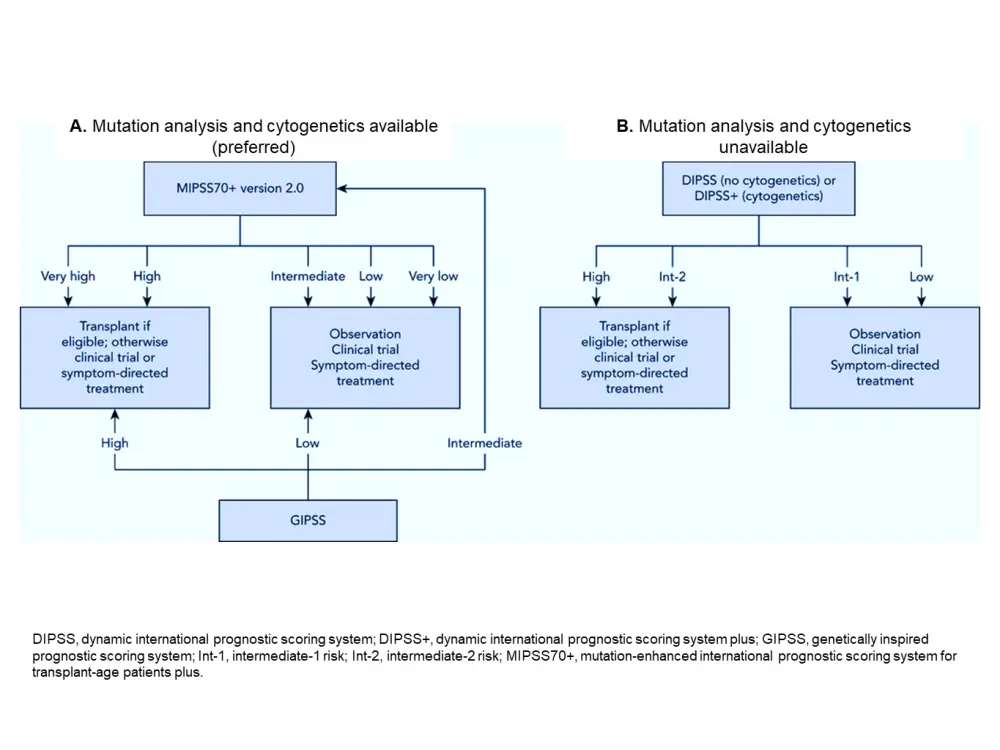
An unresolved problem is the accurate timing of HSCT. The decision of immediate or delayed HSCT may need to be discussed primarily in younger patients with good clinical status and high genetic risk (as assessed by GIPPS, for example). Here, prognostic models can provide adequate guidance for initiating bone marrow transplantation.
Special situations1
Post-ET/PV myelofibrosis
MPN management guidelines do not differentiate between therapeutic management for PMF patients versus those with post-ET/PV myelofibrosis. Using the MIPSS70 is not applicable in these cases as relevant HMRs for PMF, such as ASXL1, EZH2, and IDH1, are not predictive for post-ET/PV myelofibrosis outcomes. Therefore, the MYSEC-PM was developed explicitly for patients with post-ET/PV myelofibrosis. It identified six variables (age, Hb < 11g/dL, platelets < 150 Gpt/L, peripheral blasts ≥ 3%, constitutional symptoms, lack of CALR mutation) associated with poor outcome (Table 1) and subgroups patients into four risk levels accordingly. In a validation study in a post-ET/PV cohort, MYSEC-PM was superior to IPSS and DIPSS, allowing reclassification of patients from higher to lower risk categories. Therefore, the authors recommend using MYSEC-PM in patients with post-ET/PV myelofibrosis, although routine use needs to be further evaluated. Of note, age is a continuous variable in the MYSEC-PM and has a significant impact. This may unduly reduce the number of patients found eligible for HSCT, and it is therefore recommended to use other models (DIPSS or DIPSS+) for help with therapeutic decision making.
Prefibrotic myelofibrosis
In 2016, PMF was divided into two stages: prefibrotic and overtly fibrotic. Prefibrotic PMF patients have outcomes intermediate between ET and overtly fibrotic patients. The IPSS could discriminate low-, intermediate-, and high-risk patients. However, prefibrotic myelofibrosis patients with intermediate-risk scores had a significantly longer OS (> 10 years) than overtly fibrotic patients. Hence, prefibrotic PMF patients with intermediate risk (based on IPSS) should not undergo HSCT. However, high-risk patients have inferior OS, suggesting HSCT as a therapeutic option in this setting. MIPSS70, MIPSS70+, and MIPSS70+ v2.0 were validated in a cohort of patients with overtly and prefibrotic myelofibrosis. For patients with no available genetic data, the IPSS, DIPSS, and DIPSS+ may be useful alternatives for determining treatment intensity.
Conclusions and future directions
Research in past decades acquired enormous clinical and molecular data in PMF, leading to prognostic models with multiple variables and high data granularity. Earlier models based mostly on clinical data have evolved, and increasingly genetic information is integrated for risk assessment. However, these new models need further validation, especially when applying them to determine eligibility for HSCT.
A significant limitation of existing models is the lack of treatment options for patients with PMF. While prognostic models are critical to identify patients with high-risk features who benefit from HSCT, they have little impact on treatment decisions in transplant-ineligible patients. Due to a lack of drugs for these patients, management is similar between low- and high-risk groups and consists mainly of ruxolitinib, fedratinib, and hydroxyurea. Moreover, treatment initiation is primarily based on symptoms rather than genetic risk type. Treatment strategies are needed that consider those novel genetic markers, to maximize the utility of prognostic models.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

