All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MPN Advocates Network.
The mpn Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mpn Hub cannot guarantee the accuracy of translated content. The mpn and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The MPN Hub is an independent medical education platform, sponsored by AOP Health, GSK, Sumitomo Pharma, and supported through educational grants from Bristol Myers Squibb and Incyte. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MPN content recommended for you
Genetic testing in the diagnosis and management of patients with BCR-ABL1-negative myeloproliferative neoplasms
Your opinion matters
Are these recommendations in line with what you see in clinical practice?
Genetic and genomic testing are of increasing importance in the diagnosis and management of patients with hematologic neoplasms. Nevertheless, due to the variety of available genetic targets and limited evidence base, the appropriate use of these diagnostic tools in a clinical setting can be challenging. The British Society for Haematology (BSH) has addressed this issue via the creation of a Good Practice Paper, which has been published by Cross et al.1, to provide a consensus or a consistent approach on the use of genetic and genomic tests in patients with myeloproliferative neoplasms (MPN), myelodysplastic syndromes/myeloproliferative neoplasms (MDS/MPN), myeloid/lymphoid neoplasms with eosinophilia and rearrangements of PDGFRA, PDGFRB or FGFR1, or with PCM1-JAK2, and mastocytosis. Here, we summarize the recommendations for BCR-ABL1-negative MPN; the guidance on genetic testing in atypical MPN subtypes is summarized separately here.
Good-Practice Papers
Good-Practice Papers aim to recommend good practice in areas where the evidence is limited but a degree of consensus or uniformity may improve patient care. The Grading of Recommendations Assessment, Development and Evaluation (GRADE) nomenclature is used to indicate the quality of evidence (summarized in Table 1) and the strength of a recommendation: in Grade 1 (strong) clinicians are certain that benefits do, or do not, outweigh risks and burdens; in Grade 2 (weak) clinicians believe that benefits and risks and burdens are finely balanced, or appreciable uncertainty exists about the magnitude of benefits and risks.
Table 1. The quality of evidence*
|
(A) High |
Further research is very unlikely to change confidence in the estimate of effect |
|
(B) Moderate |
Further research is likely to have a significant impact on confidence and may change the recommendation in the estimate of effect and may change the estimate |
|
(C) Low |
Further research is very likely to have an important impact on confidence in the estimate of effect, and is likely to change the estimate |
|
(D) Very Low |
Any estimate of effect is very uncertain |
|
*Adapted from the BSH Guidelines Committee2 |
|
Methodology
A PubMed search was performed from January 2018 to September 2020 for the terms: (myeloproliferative OR polycythemia OR thrombocythemia OR myelofibrosis OR eosinophilia OR mastocytosis OR neutrophilia OR myelomonocytic OR eosinophilic CEL OR CNL OR CMML OR JMML) AND (mutation OR variant) AND (diagnosis or prognosis). The search returned a total of 135 relevant papers.
Screening investigation for erythrocytosis, thrombocytosis, suspected myelofibrosis, and atypical thrombocytosis
Molecular screening for MPN driver mutations (JAK2, CALR, and MPL) is highly likely to identify a mutation in almost all patients with polycythemia vera (PV), and 85–90% of patients with essential thrombocythemia (ET) and primary myelofibrosis (PMF). Assessment for these drivers is performed on peripheral blood DNA.
- Sequential single-target assays can be considered; however, multi-target assays, mostly next-generation sequencing (NGS), are more cost-effective.
- Broad myeloid NGS panels to screen suspected cases is not considered cost-effective; however, if larger panels are used, the initial assessment and report should only include common MPN driver mutations (Table 2).
Table 2. Targets in suspected MPN*
|
CML, chronic myeloid leukemia; ET, essential thrombocythemia; MPN, myeloproliferative neoplasms; PMF, primary myelofibrosis; PV, polycythemia vera. |
||
|
Presentation |
Variant |
Frequency |
|---|---|---|
|
Erythrocytosis |
JAK2 V617F |
96–97% PV |
|
JAK2 exon 12 mutations |
~3% PV |
|
|
Thrombocytosis |
JAK2 V617F |
50–60% ET |
|
CALR exon 9 mutation |
25–30% ET |
|
|
MPL exon 10 mutation |
3–11% ET |
|
|
BCR–ABL1 fusion |
To exclude CML |
|
|
Suspected PMF |
JAK2 V617F |
50–60% PMF |
|
CALR exon 9 mutation |
15–35% PMF |
|
|
MPL exon 10 mutation |
6–9% PMF |
|
|
Suspected CML |
BCR-ABL1 fusion |
100% CML |
- Reporting mutant allele burden is not necessary unless it is useful for prognosis.
- Reporting low allele burden (e.g., <1% JAK2 V617F) is important and a result of low levels of JAK2 V617F and the MPN phenotype should be followed by CALR and MPL screening.
- Specific CALR mutations (Type 1, 52-bp deletion; Type 2, 5-bp insertion; Type 1-like, and Type 2-like) should be reported routinely due to their prognostic value in PMF.
- There is no clinical value in routine screening for the JAK2 46/1 haplotype or polymorphisms in TERT, as these are associated with a weak predisposition to MPN.
- Clinical context is another important consideration to decide screening assays.
- JAK2, CALR, and MPL screening (Table 2) should be performed in patients with persistent erythrocytosis or thrombocytosis (Grade 1B)
- BCR-ABL1 should be excluded in thrombocytosis in the absence of MPN driver mutations or with atypical features (Grade 1B)
- JAK2 V617F screening is recommended in patients with normal blood counts and unexplained splanchnic vein thrombosis (Grade 1B) and may be considered in selected patients with unexplained cerebral vein thrombosis (Grade 2C)
- CALR screening should be considered in patients with splanchnic vein thrombosis or cerebral vein thrombosis (Grade 2C)
- JAK2, CALR and MPL variants should be assessed for patients with arterial or unprovoked venous thrombosis who have mildly/variably elevated hematocrit or platelet count that persists for 2–3 months (Grade 2C)
Testing for additional somatic driver variants with myeloid gene small variant panels +/− cytogenetic analysis
Additional driver mutations are shown to have an impact on prognosis, the risk of disease progression and implications for treatment response in MPN, read more here.
Frequencies of additional somatic driver variants are often higher in PMF or secondary MF and/or blast phase of other MPN or MDS/MPN. The minimum requirement for genes to be assessed for MPN in the National Genomic Test Directories3 include KRAS, NRAS, TP53, JAK2, CALR, MPL, ASXL1, CBL, CHEK2, CSF3R, CUX1, DNMT3A, EZH2, IDH1, IDH2, IKZF1, KIT, NFE2, SF3B1, SH2B3, SRSF2, TET2, U2AF1, HRAS, RUNX1, SETBP1, and ZRSR2 (multi-panel NGS).3 It is preferable to run these panels on bone marrow (BM) if available, but peripheral blood is also acceptable. Due to high cost and the potential to identify incidental clonal hematopoiesis, it is not considered appropriate to use panels in all MPN patients; however, these screenings may be useful in specific situations.
Patients with a suspected MPN who test negative for driver mutations
Erythrocytosis: Patients with unexplained erythrocytosis and without JAK2 V617F may be considered for a BM biopsy and JAK2 exon 12 mutation; most patients are diagnosed with idiopathic erythrocytosis if there is no secondary cause. In patients JAK2-unmutated PV, testing may be considered; however, there is no evidence to guide this approach.
Thrombocytosis or suspected PMF: Screening for additional mutations in patients with triple-negative ET and PMF may be useful in diagnosing a clonal disorder depending on age, clinical presentation, and gene panel content. Testing is recommended in patients with BM histology and clinical features consistent with PMF using myeloid gene panels combined with conventional karyotyping (or single nucleotide polymorphism assay). In patients with triple-negative ET, some patients may harbour a non-canonical mutation in JAK2 or MPL, or in another driver gene; testing for a clonal marker may be advisable for younger patients (<60 years old) with BM histology typical of ET (or MPN-unclassifiable or suspected prefibrotic MF) where confirmation of a clonal disorder may inform on disease progression with a broad panel with non-canonical variants in JAK2 and MPL and a range of other driver genes. Another case may be patients with significant thrombocytosis (>600 × 109/L) where no reactive cause has been identified, and cytoreduction would be helpful for a clonal disorder, such as unexplained thrombotic events, especially in younger patients. In triple-negative patients with thrombocytosis, screening for driver mutations may be considered at intervals (e.g., 5 years) if thrombocytosis is persistent. BM histology is still very important to confirm an MPN diagnosis in these patients.
- A myeloid gene panel and cytogenetic analysis is recommended for patients with BM histology and clinical features consistent with PMF (+/− features suggestive of MDS or MDS/MPN) who test negative for JAK2, CALR, and MPL (Grade 1B)
- A myeloid gene panel and cytogenetic analysis is not recommended for most patients with JAK2-, CALR-, and MPL-negative erythrocytosis or thrombocytosis but may be considered in individual cases (Grade 2C)
- Testing is not recommended in the event of normal or reactive BM cytology.
Figure 1 summarizes the recommended guidelines in patients with suspected MPN who test negative for driver mutations.
Figure 1. Flow of genetic testing in case of suspected ET/PV/PMF with a negative result for driver mutations*
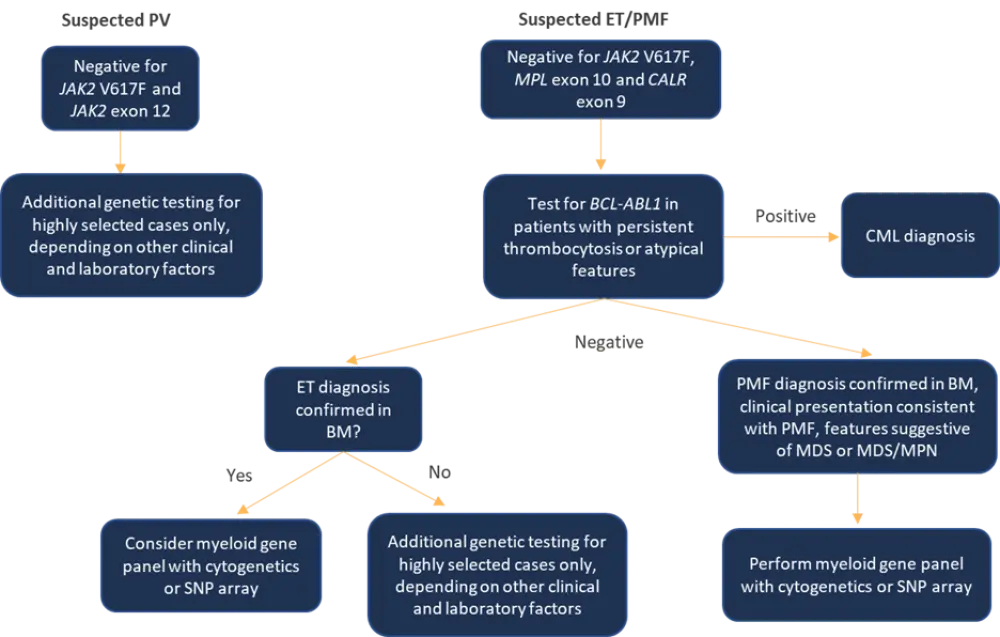
BM, bone marrow; CML, chronic myeloid leukemia; ET, essential thrombocythemia; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasms; PMF, primary myelofibrosis; PV, polycythemia vera; SNP, single nucleotide polymorphism.
*Adapted from Cross et al.1
Patients with known driver mutations
In patients with known clonal disorders with JAK2, CALR, or MPL mutations, myeloid gene panels can provide supplementary information on diagnosis and prognosis at presentation or suspected transformation with a potential to inform treatment decisions for targeted therapy in the future. These panels may also be useful to diagnose other disorders associated with JAK2 V617F, including MDS/MPN. They also represent a valuable tool for evaluating prognosis when deciding on a patient’s suitability for allogenic stem cell transplantation.
In patients with blast-phase MPN, myeloid panel testing is recommended for prognostic risk stratification, or this information would allow patients to enter a clinical trial. Repeated testing at the chronic phase is rarely helpful.
Myeloid gene panel testing is recommended:
- in patients with MPN and JAK2, CALR, and MPL mutations as well as additional cytopenias at diagnosis, unexplained ring sideroblasts or other dysplasia, increased blasts (including blastic transformation), peripheral blood monocytosis or atypical clinical features (Grade 1B)
- in all patients with PMF and secondary MF, who are candidates for allogenic stem cell transplant, combined with conventional karyotyping (Grade 1B)
- other patients if the additional genomic data will guide clinical management (Grade 2C).
- younger patients who fulfil the BSH criteria for ET, PV, or MPN-U at diagnosis with atypical clinical features requiring additional surveillance
- patients who are not candidates for allogeneic stem cell transplantation
- patients requiring testing to enter a clinical trial
Disease monitoring: Quantitative assays of clonal burden
Current research in patients receiving pegylated interferon-alpha and ruxolitinib has not yet confirmed that a particular level of molecular response is associated with a more favourable vascular or transformation risk; therefore, molecular response is not considered a treatment target. As such, there is no evidence to recommend routine quantitative monitoring of clonal burden:
- Quantitative assays of mutant allele burden are not recommended for most MPN patients but may be considered where demonstration of molecular response may influence clinical management (Grade 2C)
- High sensitivity assays of mutant allele burden are recommended following post-allogenic stem cell transplant to monitor for residual disease (Grade 1C)
Conclusion
Genetic and genomic testing provide novel insights in the diagnosis and management of hematologic disorders and have an impact on clinical practice. Considering cost-effectiveness and clinically appropriateness, it is vital to assess its utility on a case-by-case basis rather than adopting an untargeted approach.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

