All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MPN Advocates Network.
The mpn Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mpn Hub cannot guarantee the accuracy of translated content. The mpn and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The MPN Hub is an independent medical education platform, sponsored by AOP Health, GSK, Sumitomo Pharma, and supported through educational grants from Bristol Myers Squibb and Incyte. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MPN content recommended for you
Prognostic value of ASXL1mut in patients with PMF and SMF
Do you know... The NGS prognostic model for patients with myelofibrosis was recently proposed by Luque Paz, et al. It includes four genomic groups. Which of the following groups is not included in the model?
The underlying molecular architecture in myelofibrosis (MF), whether primary or secondary, is still incompletely understood. Multiple gene mutations, including ASXL1 mutations (ASXL1mut), have been identified as conferring a high risk of poor outcomes in myeloid malignancies.1 However, the value of ASXL1mut as a prognostic factor in MF has been recently questioned, and a novel model, named “next-generation sequencing (NGS) with a custom RNA-baits panel” was put forward.2 This model includes four categories: patients with TP53mut; high-risk patients (≥1 mutation in EZH2, CBL, U2AF1, SRSF2, IDH1, IDH2, NRAS, or KRAS); patients with ASXL1mut-only; and other patients.2
In the article by Guglielmelli, et al.1 published in Blood Advances, the prognostic role of ASXL1mut was reviewed, paying particular attention to the difference between patients with primary MF (PMF) or secondary MF (SMF). We summarize the key results below.
Study design
To assess the value of ASXL1mut as a prognostic factor, 523 patients with MF were analyzed: 63% with PMF and 37% with SMF. Of the SMF patients, 44% had polycythemia vera-MF, and 56% had essential thrombocythemia-MF. NGS was used for mutational analysis.
Results
The baseline characteristics for patients with PMF and SMF are shown in Table 1. Patients were followed for a medium of 81 months in the PMF group and 77 months in the SMF group. For the whole cohort, the median age at diagnosis was 60 years (range, 18−90) and 60% of patients included were male.
Overall, 62% of patients had JAK2mut, 24% had CALRmut, 5% had MPLmut, 8% were triple-negative, and 2% double-mutated. There were significantly more patients with PMF who were triple-negative compared with patients with SMF (p < 0.0001).
Table 1. Baseline characteristics*
|
BM, bone marrow; Hb, hemoglobin; PB, peripheral blasts; PMF, primary myelofibrosis; SMF, secondary myelofibrosis. |
|||
|
Variables, % (unless otherwise specified) |
PMF |
SMF |
p value |
|---|---|---|---|
|
Male sex |
62 |
60 |
0.09 |
|
Age at diagnosis, years, median (range) |
58 (18−90) |
63 (20−87) |
0.0037 |
|
Hb g/dL, median (range) |
12.3 (3.8−17.5) |
11.5 (5.9−17.5) |
0.0087 |
|
Platelets, × 109/L, median (range) |
413 (10−1,800) |
339 (14−1,660) |
0.0047 |
|
PB, %, median (range) |
0 (0–16) |
0 (0–18) |
0.0014 |
|
PB ≥1% |
18 |
31 |
0.0003 |
|
BM fibrosis Grade ≥2, median (range) |
153 (49) |
174 (97) |
<0.0001 |
|
Constitutional symptoms |
41 |
57 |
0.0007 |
The prevalence of driver mutations and myeloid malignancy-associated mutations is shown in Figure 1. An equal percentage (30%) of ASXL1mut was recorded for both the PMF and SMF groups.
Figure 1. Mutation prevalence in patients with MF*
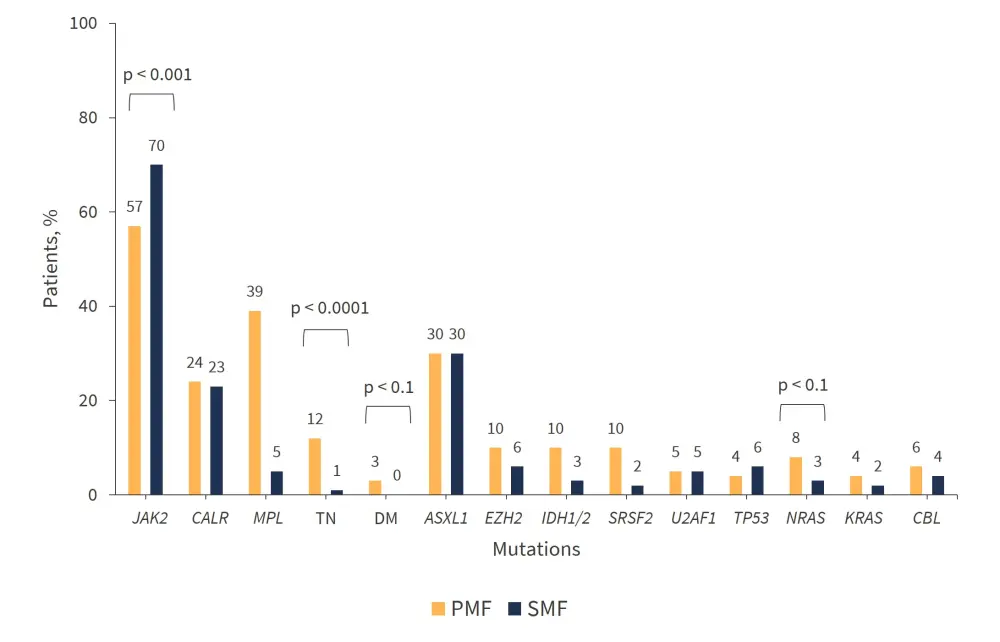
DM, double mutated; PMF, primary myelofibrosis; SMF, secondary myelofibrosis; TN, triple-negative.
*Adapted from Guglielmelli, et al.1
For patients with PMF, ASXL1mut was significantly associated with characteristics that indicate a higher-risk disease, such as:
- Older age (median, 64 years vs 56 years; p < 0.0001)
- Male sex (74% vs 26%; p = 0.0042)
- Increased leukocyte count (11.9 × 109/L vs 8.3 × 109/L; p = 0.0083)
- Decreased hemoglobin levels (11.2 g/dL vs 12.7 g/dL; p < 0.0001)
- Reduced platelets (252 × 109/L vs 517 × 109/L; p < 0.0001)
- Increased peripheral blasts (1% vs 0%; p < 0.0001)
- Bone marrow fibrosis Grade ≥2 (69% vs 40%; p < 0.0001)
- Constitutional symptoms (57% vs 34%; p = 0.0001)
- Transfusion dependence (43% vs 20%; p < 0.0001)
ASXL1mut was found to significantly cluster with EZH2mut (p < 0.0001), SRSF2mut (p < 0.0001), U2AF1mut (p = 0.0002), CBLmut (p = 0.0006), NRASmut (p < 0.0001), KRASmut (p = 0.0051), RUNX1mut (p = 0.0158), and SETBP1mut (p < 0.0001).
Only four mutations were significantly associated in patients with SMF: MPLmut (p = 0.0207); EZH2mut (p < 0.0001); U2AF1mut (p = 0.0301); and NRASmut (p = 0.0122). Compared with patients with PMF, the variant allele frequency (VAF) of ASXL1mut was decreased in patients with SMF (42% versus 26%; p = 0.0129).
NGS model
With respect to the NGS model categories, the prevalence of mutations of interest are shown in Table 2. Patients with ASXL1mut and TP53mut were more frequently diagnosed with SMF compared with patients in the high-risk, or other categories (44% and 48% versus 28% and 38%, respectively). Patients in the high-risk group were enriched for triple-negative, and CALRmut was found more often in patients in the ASXL1mut-only group, and other categories, compared with the TP53mut and high-risk groups (25% and 27% versus 12% and 18%, respectively).
Overall survival (OS) was lowest in the TP53mut and high-risk groups (p = 0.0039). Patients with ASXL1mut-only showed inferior OS compared with patients in the other categories (p = 0.0118) (Table 2).
Table 2. Patient categories according to the NGS model and univariate analysis of OS*
|
NR, not reached; OS, overall survival; PMF, primary myelofibrosis; SMF, secondary myelofibrosis. |
||||
|
NGS category, % |
PMF |
SMF |
p-value |
Median OS, |
|---|---|---|---|---|
|
TP53mut |
4 |
6 |
0.46 |
38 (14−110) |
|
High-risk |
30 |
20 |
0.0097 |
55 (45−85) |
|
ASXL1mut-only |
10 |
16 |
0.0412 |
124 (91−156) |
|
Others |
56 |
59 |
0.53 |
193 (142−NR) |
PMF
When analyzing the OS in patients with PMF separately, the TP53mut and high-risk categories were associated with the poorest OS with a median of 58 months (range, 20–126) and 55 months (range, 36–85), respectively, although the difference was not significant. In patients with PMF and ASXL1mut-only, the OS was significantly decreased compared with patients in the “others” category, with a median of 103 months (range, 78–not reached [NR]) versus 320 months (range, 178–NR) (p = 0.0170).
In patients in the high-risk group, 62% carried ASXL1mut, which was associated with a decreased OS compared with non-carriers, with a median of 47 months (range, 31−73) versus 102 months (range, 34−317) (p = 0.0240). The VAF was significantly increased in patients in the TP53mut group, and the high-risk group, compared with the ASXL1mut-only group (47% versus 34%; p = 0.0303).
SMF
When analyzing the OS in patients with SMF separately, it was observed that the TP53mut category was associated with the poorest OS; however, the three other categories did not have significantly different OS:
- in the ASXL1mut-only category, the OS had a median of 141 months (range, 56–171).
- in the “others” category, the OS had a median of 131 months (range, 106–NR; p = 0.5188).
- in the high-risk category, the OS had a median of 58 months (range, 45–174; p = 0.3606).
- ASXL1mut occurred in 63% of patients in the high-risk category but did not significantly affect the OS (p = 0.3296).
In patients with SMF, the VAF was not significantly different in the TP53mut and high-risk categories compared with the ASXL1mut-only group.
For patients with PMF, the High-Molecular Risk-Dynamic International Prognostic Scoring System (HMR-DIPSS) was the best at predicting death at all time points compared with the NGS-DIPSS combination. The Mutation-Enhanced International Prognostic Score System (MIPSS70), and the MIPSS70+ version 2.0, include ASXL1mut in the scoring system, and achieved the highest values for performance and accuracy.
In patients with SMF, the NGS classification system was superior to the HMR classification, and the highest values for performance and accuracy came from its integration with the MYelofibrosis SECondary to polycythemia vera and essential thrombocythemia-Prognostic Model (MYSEC-PM). The MIPSS70 and MIPSS70+ version 2.0 classifications were mostly inferior compared to other prognostic models for patients with PMF.
In PMF, ASXL1mut was associated with high-risk features, unlike SMF. Even in the absence of any other high-risk mutations, the presence of ASXL1mut confers a negative prognostic risk for patients with PMF.
Conclusion
To the best of the authors knowledge, this study is the first to highlight the differential prognostic impact of ASXL1mut in patients with PMF compared with SMF. The value of scoring systems that include ASXL1mut, such as the MIPSS70 classification, for patients with PMF was also highlighted. This difference between PMF and SMF with regard to their molecular architecture, strengthens the argument that they are two distinct biological entities and require specific prognostic models for each.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content



